Available ZH-attP lines and their genomic location of the attP landing site
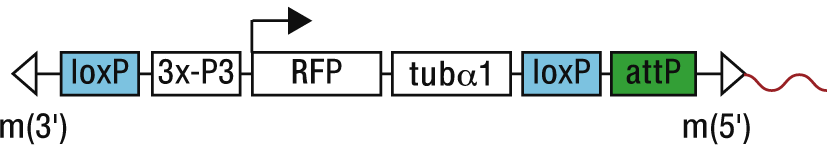
For an exact analysis of the location of an attP landing site, or the distances to neighboring genes and the like, we provide купить квартиру в новостройке here the flanking sequences of 25 attP lines. The given genomic sequences (5'-3' direction) immediately flank the landing site at the mariner 5' terminal repeat (m5'), which lies at the 3' end of the landing site construct (indicated in red, Figure below).
| ZH-attP line |
chrom. 1 |
|
| zh-1E |
X 1E |
|
| zh-2A |
X 2A |
sequence |
| TATATATTTC TTCGCGGTAA CCTCTGCCCA TTTTCGGATC TTTGTGCTGG CTCGCTCATC CATCCCTACC TACCGACTCC AAGCGACTAC TACTCCTTCC CTGGTCAGGC CTAGCCATGT GACTACTTCA TCCGCAAGTA AGCGATACCT CGGCTGGGCT CTTGTTTGTA ACAATCTTAC TTTCTGCATG CATTTGCTGT ACTAATGATT ATATAGAGCA CACACGGGCC CACACACACC ACACCAAGTC |
| zh-3Aa |
X 3A |
|
| zh-3Ab |
X 3A |
|
| zh-3B |
X 3B |
sequence |
| TAAGGTCGGG TCTCCAAAAA AGGCATAAAA CCCAGAAAAG GCAATGAAAA TTGCCAAGTC GCATTATTTA TCGTATACGC ATCGGGGAGT ATAGCGAGCA CACACACACA CGCACACAAG TGCTCTTAAA TGCACACACA CAGACACAGA TACACACACA CACACATGCA GTGTTAGAGC GGAAGGGGGC AAAACAAAAT GGCGCTGCAT TCATAATGGC GGTTGGTGGT TCGGTCGCTT CCTGTCGTCG |
| zh-5D |
X 5D |
|
| zh-6E |
X 6E |
sequence |
| TAATCGTTGG TCACCTCCTG GAATTCCTGT TCGCCTGGTA ATTTAAGCCG AAAATCTCTA AATTTTGCTT ACAAAATGAG CATTTTAGTG GCAAATTTTG GGATGTTTGG TACCAGGCCA TCATACTCAG TAGCGGGTGT CTACTGAAAA CCAACTAATC GTTGGTCGCC TCCTGGAATT CCTGTTCGCC TGGTAATTTA AGCCGAAAAT CTCTAAATTT TGCTTACAAA ATGAGCATTT TAGTGGCAAA |
| zh-14F |
X 14F |
|
| zh-20C |
X 20C |
sequence |
| TATATCTCCG CCTGTCTAAG CCTTAAGTCA GCTCATTTCC TAAATGCTCA CTAATCATCC AAACTACCGG CATTCAAAGA CGACGACCGC CTTCGCACAT CTCAATTAGT GGCAATAAAT TTAAGTTTCA TTTAGGTGCG AAGCACAGTT |
| zh-X |
X-linked |
|
| ZH-attP line |
chrom. 2 |
|
| zh-21F |
2L 21F |
|
| zh-22A |
2L 22A |
sequence |
| TACATTGTTT ATACGAGAGC TGTCTTTGCA CGGCTCCCAA AAATATGACT CACAAGAAGT TTGACTTCAC TCCTTCATTC CGAGGTATGA AATATTTATA AACGCATAAT GCATGGCATG TGAAGTGAGG TGAGATGAGG TGACGTTTGA AGGATGCACT GCGAAGCAGC CAGCTGCAAG ATCATCATCA AATGCATCAA CCCCTCAACG GCGAACCCAT CGACGACTTC AAACGAAAAC GAAGCGTGTT |
| zh-27F |
2L 27F |
|
| zh-30A |
2L 30A |
sequence |
| TAGATGGTGG TGTCCTTTGA AATGTTGTTG CAAATTGTTA GCTCTTGTCC CATAAAAGTG CACATAAAAT CAAAAAGGGG AAAAATGTGC GGGCCTCCCT CCCCTAACAA TTCCCAAACA TTTTCGCATG GCACGCCAAG CAATTAAAGC GGCCCATACA AAACTCCAAC TCCGCGTTGC ATGTGGCAAA CTTCTGGTTC CCAACTGGCG CCCAAATGGC GGACATGTCG ACGGCGGCGC CTCAATGTCA |
| zh-30B |
2L 30B |
|
| zh-32C |
2L 32C |
|
| zh-35B |
2L 35B |
sequence |
| TATTAAGTGG CTCTTTAGGA CCTGTTAAAA ATGCCCGAAC ACGCCCGCCC AAAAAAGTTG GCATTGCTAA CCCAATTTTC CCCGGTATGG GAATCTGCGT GCGATTCAAA TTTCCATTTC CTTCAATCGC CATTGGCCAG CTCGGCCAAA CACACGCACA CCTGCCGAGC GGGGCCAACA AATTTCACTT ATTGCCAATT TGCTGGCAAT GAAATAAAAT ACTTTTCTTC CGCGACGCGC CTGTTTGTTG |
| zh-36B |
2L 36B |
sequence |
| TAAAACAAAA GTAAGTTGAG TAGCCTCGGT ACTTCGAGAA GATTTCCCAA TCTAAGCGCC ACTTTGGACA TTCGATACCC ATTTCGTTAT TAGTTTTCTG CTGAAATGGA ATTTTTCTGC AGTATATAAT GTGGCATTCG CCCCATAAAT TGTCAACGCA TGTTGTCAAG GTGCTTCGGA CACTTTTTTC AATTAATTGG CGAAACTTCA AATTTCACTT TTTCTTTATT TCTTGCTTAT TTTTTTTTCT |
| zh-38D |
2L 38D |
|
| zh-44F |
2R 44F |
|
| zh-45A |
2R 45A |
|
| zh-46A |
2R 46A |
|
| zh-46D |
2R 46D |
|
| zh-49B |
2R 49B |
|
| zh-49D |
2R 49D |
|
| zh-50C |
2R 50C |
|
| zh-51C |
2R 51C |
sequence |
| TAAGTGGCAT TTTTAAATTG ATTAAAAAAG AATTGGTGAT GCAGCCAGTG CCTTCTTCGC AGTTTTCATT CGCTACTCCA GCTCATTGTG GCTCTGAATT TTCCCCTTGT TTTTCGCTGG CGTTTAATTA ACGAGGCCTG GACGACATTT GTGCCGAGCG ACATTTTTAA TTTCCTTTAT TTTGTACTCA GCAACATCGG AAGTGCCGTC AGCCAAACCG GAAGTACGCC GCTCACACAC ACACTCACAC |
| zh-51D |
2R 51D |
sequence |
| TAAAAGGGTG GGATGTGACG TTAGCTCCTC AACTACCTGA AATTGTTTTA CTAATTTGAT AAGTAACAAG GAACTGTTGC AGCTATTTTC TTTATTTCTT TAATTTACTA AATAAAACAT ATATTCTCTT AATCATAGCA TTTTCACATT GTTAAAAGCT AAGTTTTTGA AAGCCGCTCG CCTGCATGAT GCCACTATGC CCAATTCGAT CCACTGTGCA ATATCTTGTG AACTTGATTT GGGGAGCGAA |
| zh-52E |
2R 52E |
|
| zh-58A |
2R 58A |
sequence |
| TAGATATTAA CAGCAAAATG CAATTAACTA GCACCATGAC CGTTCGAAAA ATTCGCGACA ATTTAGCGAC GCTGTGGAAA TGCAAACCTC TACTGCCAAG AATTCGTGGT CCGCCGCATG ACAACGGAAA TTAAATTGCA TGAAATTGGA AAATTATTAC ACGCAGGCAC ACAGAGAGAG TGGAAAATGC ATTTGCATAG TGCTGAGGTA GCCATGCAAT TTGGCCGGCC ATTTAAGCCA AATTAAGGCC |
| zh-59D |
2R 59D |
|
| zh-60E |
2R 60E |
|
| ZH-attP line |
chrom. 3 |
|
| zh-62B |
3L 62B |
sequence |
| TATGGTAATA AACACTCACC ATGGGAGTTA ATATTTGTAT TCTTTACATA TTTTCAGAAA TAACATTATC TGTATGACTA CGCACGGAGC CTCGAAATTG ACTAAATGGC TTCAGAGGCT GTGGCCCTTA GTTTGCTCAA TTTCAGATTT CTTCTTGAAA AGTCAGTAGC GGTCTTAGGG TTCCAAATAT AATGAAAAAG TTAAGGGGAT AGAGTTCAAA AGCAATTTTC TATTGTTCGT GCTTAAAAGT |
| zh-64A |
3L 64A |
sequence |
| TAATGCAGCA ATGTTGCCCA AAAGTTGCCA TTGCAGTTGG GAAATCTTGT TGGTCGGCAA GCACGCCGTG CACCACCGCC CTATATCCTG CACGGTGAAA AAGGAAGCGA GGGGACAAAT TTTAAGTTTC ATAACAATTA ATGTATTTAA AAATAACAGG TTTGTAGAGT GAAACCGAAA GTAAATGGTT ATAAAGGTGT TTAGATAAAG GTGTTTCTCA TTCATTGATA AATCCTTCGC TTAGCCGCCT |
| zh-67C |
3L 67C |
|
| zh-68E |
3L 68E |
sequence |
| TATATATACC ATTCTTTCAG CTGGATATTC TTGTACTAAG TTGATATGTA TATTTATTTA TCAGAAACTG TTTCCATTAA AACTTGCTCC TAAAAAAATT ATTAAGTCGA TTGTGATGTG GCATCTATTG CAACACTTCC GGCTAAATTA AACGGATGAG GTTTCCTGGA GTGTAAAGCA AGGCAAACAT GACGTATGGA AAATATCGGT TCACGCTCGT AAATCGCCCC AAAAACACGC TCCATGCACA |
| zh-70C |
3L 70C |
|
| zh-75C |
3L 75C |
sequence |
| TATATATCCG CACATGCAAC CCTCAGCTAC CTTCAACTAC ATTTTGTATC TTTTGTTTTC TCGGATGTTG CGTATTTTGA CAATGTGCCT ACAGTTTGCT TTGTGTGTCT TTTGTTCTGC GAAGGGTTTT CTCCTGGGTC TATAAAATTC GCAACAAAGT CAATTTCAGA ACTATCGGCA TACCGATTCC AGTGGATCTT AGAAAATTGT GTTTTTGTAA AAGGATTAGT TATGGTTATC CTGTTTCCTG |
| zh-75D |
3L 75D |
|
| zh-82F |
3R 82F |
|
| zh-83A |
3R 83A |
|
| zh-83D |
3R 83D |
sequence |
| TAGATGATTG TATGATTAGG AACAAAAACA GGCGCCCAGG AAATTAACCA ACATTTGGGC AGCTAATTGG CTGCCTAAAT GGCTCAAGCT GCTCGTGTGG AAATCCATTG AGGGTCCTGC TCCGAATCCA ATGAACCGCA CAGCGTAAAG TGTCGTCTTC CACACTTCAT TGAAGGCTGT CAAAAAATTT TCAATTTACC AAATTAGTCC GGCAGATGTG TGGGCTCGAA TACGGAAGGA TCCCAGGACA |
| zh-85D |
3R 85D |
|
| zh-85Ea |
3R 85E |
|
| zh-85Eb |
3R 85E |
|
| zh-85F |
3R 85F |
|
| zh-86Da |
3R 86D |
sequence |
| TAATCAACTT CAGGCTGGCA GGACTCATTA AAAATTGAAA TCATTTTCGG GTGCACGGCG AAAAGTCAAC GCAAATTGCA GCCTATTTGT CAGTCGGCAT GCGTTTTAAC CTCGCTTTTC CTATCTTTTT GACTCGCAAC GTGGGCGTGG CTGTCATTTT AATTAGGGCA AGAAATGAAA AACATGGGTC ACAAACAAAA AAACGCAAAA AAACCAACAC ACCAAAAGGA AAGTTTGCCA ACAGCCTGAG |
| zh-86Db |
3R 86D |
|
| zh-86Dc |
3R 86D |
sequence |
| TATATGGGAG ATTATATAAA GGGTTAAACG AATTAATATG GGTACTTGGA TCAACACTAC ATTTAAACCT ATTAAATGCA CCAAATACCG ATTTTTATAA TAAGCGAAGT ATTTTTTGTT CAAAAGTAGG TGGGAATCCG ATTGAAGTGT AGCAAGTGGA AGCCCCCACA AAAAAATGTT GGCCCTTGCA TATTGTTTTC CGTTGTTTGG GAGCCCAAAA AAGCTGGATT CGCGAGCAGA AATCGTGATG |
| zh-86Fa |
3R 86F |
sequence |
| TATACAAGGT TTTGCAATAT GTCGCATTGC TATGTATTCT TTTGGATCTG TTAAATCGTT TACTTTATCG TTAATCGTTA ATCTCAAAAT GCTTTGTATT TGAACAGTAA ATTACGTTGT CACTTGCTCG AAGTGTTTGC ACTTTATTTC GGTTATTGTT TTATGCCTTT ATTCCTATTC GGTTTTCATA TGCTCTCTCT TTTGTATTTA TATATAAAAA TATCCTTTTA TACAGTTGTG TTTTCGGTTA |
| zh-86Fb |
3R 86F |
sequence |
| TAATAGGTCA TAGAGATATA CTTTCTTGCT ATGGTCATAA AGTTACTAAG CCAAATTTAT ATTTCTTTAT AGCCAGTTAT TTACACTTTG GCCCTAACTC TTTATCTTTT TGCCATGGAT ATAAGAAATT TACGGTCACA TGCACCCAGA AGCTAAGCAC GATATTTACT TAAAGATTTT GTTCAATTTT TAATTTCGTT CGGCTGCTTC TGTCCCCCAT CCTGAATAAT TTCTAACCGC GCCTCAGGTT |
| zh-87E |
3R 87E |
|
| zh-88A |
3R 88A |
sequence |
| TAAATTCCAT AGAACAAACT CGGGGGAAAC AAACCCTCAT AAAGTGACAA TGGCAAAAGC AATTCAATTT GAAAATTTCG ATTTAATTGT CCTTGTGTTT TCGCTGCATT GTGTACGATA TTT |
| zh-88D |
3R 88D |
|
| zh-89E |
3R 89E |
|
| zh-91B |
3R 91B |
|
| zh-92Aa |
3R 92A |
sequence |
| TAAGCCTTTA ATTGCACAGA CCGCAAACAA TGAATGGAAA AAGGGAGCTT CAACTCACCA GAAACGGACA TAAATCACGT TGAATTCGAA GAGCTGAAGA GTTGGTAGAG GGAAATCAAA ATTCTATTAA TCAGCAACGG CGGCGTTTAG CCCAAAAACA AGCGAATGAA AAAAAATTAA CACCCAACTA ATGCCGACTG CCGACAAAAG ATGACGAACC GAAATCCCGA AACAAGCTGA AATAAACCCT |
| zh-92Ab |
3R 92A |
|
| zh-93D |
3R 93D |
|
| zh-94D |
3R 94D |
sequence |
| TAAGAACTTC TAATCCTATG ACCAAAGAGT ACTCATAAGT CATTAAAGAG TACCCCCTTT TCTCTCAGTG CACCATCCAG CATTGATGGG TAACTCCTGC TCTGGAAACG AAAGTCGAAT GCATTTGGCC AAAGCCTTTG CACTGTATTG GCTCCTTTAT GCACATTCAA TTGGTGGTGT GGCTAATCTA GCGAAATTCC AGCCAACTGA GAAGGACCAA ACGACTAAAT CGAATCGCAG CTTATTTGAG |
| zh-96E |
3R 96E |
sequence |
| TACATTTTAT TTTCATGGCA AACGTGAGCC GTGCAGCACG TTAATCGATT CAAATCGATG CGACTGCCGA AATCCGCTCA CAATATGCCG ACATTTCGC |
| ZH-attP line |
chrom. 4 |
|
| zh-102D |
4 -102D |
sequence |
| TAAGTAGGTA CGCAAGAAAA ATCAAATATC ATGAGTGCAA AATTTAATGG TTATATGCAT TTGTTGCAGC CCAGACACTA CGCTAAATAA TATAAATTAA GGTATTTTCA CACATTCCAG GAAAATCAGA TTCGTCACCT TTACAATGCG CCATGAAATT TTTGGCACAC GCAATAGGAA TGACAAAAAG GCGATTCAAG AATATGCCCC TTTGAAGAAG CCCAGCACAC TGGATGCCGG CACTACAATA |
The names of the ZH-attP lines indicate the cytological position of the attP landing site. In cases where we have more than one line for the same cytological region we distinguish the lines by adding an "a" or "b" etc. at the end. ZH-attP-X could be mapped to the X-chromosome, but due to three X-chromosomal hits not to a specific cytological region yet. The lines provided with a sequence contain a putative intergenically located landing site (according annotated genes or GCs provided in Ensembl v41). Recently, Venken et al. (2006) also generated a considerable number of attP sites, which are marked by yellow, with a P-element strategy.
Targeting frequencies of various attP sites
The targeting frequencies found in the following table are based on doubly heterozygous flies, with one exception (you will find the details to the injection setup below the table).
| ZH-attP line |
integration frequency (%) |
Eye color |
| ZH-attP-2A |
0 |
Orange |
| ZH-attP-22A |
39 |
Light orange |
| ZH-attP-30A† |
41 |
Orange |
| ZH-attP-35B |
40 |
Light orange |
| ZH-attP-36B |
57 |
Orange |
| ZH-attP-51C |
42 |
Light orange |
| ZH-attP-51D |
60 |
Red |
| ZH-attP-58A |
49 |
Light orange |
| ZH-attP-62B |
39 |
Light orange |
| ZH-attP-64A |
54 |
Orange |
| ZH-attP-68E |
37 |
Light orange |
| ZH-attP-75C |
37 |
Light orange |
| ZH-attP-86Fa |
28 |
Red |
| ZH-attP-86Fb |
43 |
Orange |
| ZH-attP-96E |
44 |
Orange |
| ZH-attP-102D |
45 |
Red |
Females homozygous for the vas-phiC31 construct, located at 102D on the fourth chromosome, were crossed to males homozygous (or hemizygous) for an indicated attP site, and the offspring were injected with pUAS-lacZattB (215 ng/µl). Since line ZH-attP-2A has the attP site on the X chromosome, G0 males of this line are devoid of an attP site and therefore are not expected to give rise to transgenic offspring (G0 females of this line gave transgenic offspring).
Eye colors were determined three days after eclosion (heterozgous state). All flies transgenic for the injected plasmid reveal a reddish punctate color at the inner side of each of the three ocelli; this additional phenotype served as a convenient confirmation for transgenic flies in the cases of light orange eye color. "Light orange" is in some cases hard to detect and it is helpful to screen for transformants of such attP lines only a few days after eclosion.
Landing site ZH-attP-30A, though homozygous viable on its own, turned out to be homozygous lethal upon integration of UAS-lacZattB.
